
| Abbreviation ..................................................... | : | lac-D |
| Name ..................................................... | : | D-Lactate, etc.(R)-Lactate, D-Lactic acid, D-2-Hydroxypropanoic acid, D-2-Hydroxypropionic acid |
| Model ID ..................................................... | : | C00256 |
| KEGG ID (?) Compounds in KEGG database that match this compound. ..................................................... | : |
C00256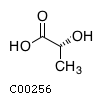
|
| Formula ..................................................... | : | C3H5O3 |
| Charge ..................................................... | : | -1 |
| Localization ..................................................... | : | Mitochondria, Cytosol, Extracellular |
| BiGG matches (?) Compounds in BiGG database that match this compound. ..................................................... | : | lac-D |
| Other associations ..................................................... | : | See reaction table below (6 reactions in total) |
|
|
||
|
In WormPaths
(?)
Pathway maps where this metabolite is represented are listed here. Letters after map name indicate the compartments the metabolite is placed in the map: (c) Cytosol (m) Mitochondria (e) Extracellular space ..................................................... |
: | Methylglyoxal detoxification (c,e,m), Pyruvate metabolism (m) |
 |
||||||
 |
||||||
To be determined |
 |
(2) acgamName: N-Acetyl-D-glucosamine Formula: C8H15NO6 Charge: 0 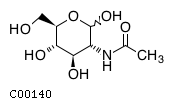 click to see details on this item + lac-DName: D-Lactate Formula: C3H5O3 Charge: -1  click to see details on this item + glu-LName: L-Glutamate Formula: C5H8NO4 Charge: -1 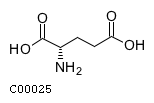 click to see details on this item + (3) ala-LName: L-Alanine Formula: C3H7NO2 Charge: 0 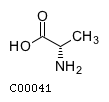 click to see details on this item + lys-LName: L-Lysine Formula: C6H15N2O2 Charge: 1  click to see details on this item + co2Name: CO2 Formula: CO2 Charge: 0  click to see details on this item |
Not determined |
|||
Not applicable |
 |
Not determined |
||||
Not applicable |
 |
Not determined |
||||
Not applicable |
 |
Not applicable |