
| Abbreviation ..................................................... | : | dgdp |
| Name ..................................................... | : | dGDP, etc.2'-Deoxyguanosine 5'-diphosphate |
| Model ID ..................................................... | : | C00361 |
| KEGG ID (?) Compounds in KEGG database that match this compound. ..................................................... | : |
C00361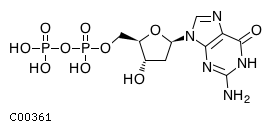
|
| Formula ..................................................... | : | C10H12N5O10P2 |
| Charge ..................................................... | : | -3 |
| Localization ..................................................... | : | Mitochondria, Cytosol |
| BiGG matches (?) Compounds in BiGG database that match this compound. ..................................................... | : | dgdp |
| Other associations ..................................................... | : | See reaction table below (17 reactions in total) |
|
|
||
|
In WormPaths
(?)
Pathway maps where this metabolite is represented are listed here. Letters after map name indicate the compartments the metabolite is placed in the map: (c) Cytosol (m) Mitochondria (e) Extracellular space ..................................................... |
: | Purine metabolism (c) |
 |
||||||
 |
||||||
 |
||||||
Not applicable |
 |
Not determined |
||||
Not applicable |
 |
Not determined |
||||
Not applicable |
 |
Not determined |
||||
Not applicable |
 |
Not determined |
||||
Not applicable |
 |
Not determined |
||||
Not applicable |
 |
Not determined |
||||
Not applicable |
 |
Not determined |
||||
Not applicable |
 |
Not determined |
||||
Not applicable |
 |
Not determined |
||||
Not applicable |
 |
Not determined |
||||
Not applicable |
 |
Not determined |
||||
Not applicable |
 |
Not determined |
||||
Not applicable |
 |
Not determined |
||||
Not applicable |
 |
Not determined |